voi.remote for Value of Information calculation - based on Chris Jackson’s VoI package
Eric Yu, Yihang Xu
2021-09-08
voi_remote.RmdThis document is heavily based on the VoI vignette written by Christopher Jackson, and is meant to show the compatiblity of the voi.remote packae with the original voi package. The voi.remote package has the exact same functionalities as voi package, but allows users to run all the VoI functions on the server rather than running locally.
Package installation
The development version of voi.remote can be installed from GitHub with:
install.packages("remotes")
remotes::install_github("resplab/voi.remote")Specifying model inputs (same as voi package)
The inputs should be a data frame with one column per parameter and one row per random sample.
set.seed(1)
nsam <- 10000
inputs <- data.frame(p1 = rnorm(nsam, 1, 1),
p2 = rnorm(nsam, 0, 2))Specifying model outputs
outputs_nb <- data.frame(t1 = 0,
t2 = inputs$p1 - inputs$p2)
outputs_cea <- list(
e = data.frame(t1 = 0, t2 = inputs$p1),
c = data.frame(t1 = 0, t2 = inputs$p2),
k = c(1, 2, 3)
)Expected value of perfect information
Computation using random sampling
decision_current <- 2
nb_current <- 1
decision_perfect <- ifelse(outputs_nb$t2 < 0, 1, 2)
nb_perfect <- ifelse(decision_perfect == 1, 0, outputs_nb$t2)
(evpi1 <- mean(nb_perfect) - nb_current)## [1] 0.4778316
opp_loss <- nb_perfect - nb_current
mean(opp_loss)## [1] 0.4778316
Using the voi.remote package to calculate EVPI
The voi.remote package contains a simple function evpi.remote to compute the EVPI using the above procedure. The function called the corresponding function in the voi package on the server.
library(voi.remote)
evpi.remote(outputs_nb)## Calling server at https://prism.peermodelsnetwork.com/route/voi/run## [,1]
## [1,] 0.476
evpi.remote(outputs_cea)## Calling server at https://prism.peermodelsnetwork.com/route/voi/run## $k
## [1] 1 2 3
##
## $evpi
## [1] 0.4760 0.4028 0.4174Analytic computation
mean_truncnorm <- function(mu, sig, lower=-Inf, upper=Inf){
a <- (lower-mu)/sig
b <- (upper-mu)/sig
mu + sig * (dnorm(a) - dnorm(b)) / (pnorm(b) - pnorm(a))
}
enb_correct <- mean_truncnorm(1, sqrt(5), lower=0)
mean_nb_perfect <- enb_correct * prob_correct
(evpi_exact <- mean_nb_perfect - nb_current)## [1] 0.4798107Expected value of partial perfect information
The function evppi.remote can be used to compute this, similar to the evpi function (which is called on the server).
Invoking the evppi.remote function.
(a) As a vector.
evppi.remote(outputs_nb, inputs, pars=c("p1","p2"))## Calling server at https://prism.peermodelsnetwork.com/route/voi/run## $pars
## [1] "p1,p2"
##
## $evppi
## [1] 0.476(b) As a list.
evppi.remote(outputs_nb, inputs, pars=list("p1",c("p1","p2")))## Calling server at https://prism.peermodelsnetwork.com/route/voi/run## $pars
## [1] "p1" "p1,p2"
##
## $evppi
## [1] 0.0822 0.4760
evppi.remote(outputs_cea, inputs, pars=list("p1",c("p1","p2")))## Calling server at https://prism.peermodelsnetwork.com/route/voi/run## $pars
## [1] "p1" "p1" "p1" "p1,p2" "p1,p2" "p1,p2"
##
## $k
## [1] 1 2 3 1 2 3
##
## $evppi
## [1] 0.0822 0.1706 0.2590 0.4760 0.4028 0.4174The evppi.remote function returns a data frame with columns indicating the parameter (or parameters), and the corresponding EVPPI. If the outputs are in cost-effectiveness analysis format, then a separate column is returned indicating the willingness-to-pay.
evppi.remote(outputs_cea, inputs, pars=list("p1",c("p1","p2")))## Calling server at https://prism.peermodelsnetwork.com/route/voi/run## $pars
## [1] "p1" "p1" "p1" "p1,p2" "p1,p2" "p1,p2"
##
## $k
## [1] 1 2 3 1 2 3
##
## $evppi
## [1] 0.0822 0.1706 0.2590 0.4760 0.4028 0.4174Changing the default calculation method
Gaussian process regression
evppi.remote(outputs_nb, inputs, pars="p1", method="gp", nsim=1000)## Calling server at https://prism.peermodelsnetwork.com/route/voi/run## $pars
## [1] "p1"
##
## $evppi
## [1] 0.0883Multivariate adaptive regression splines
evppi.remote(outputs_nb, inputs, pars="p1", method="earth")## Calling server at https://prism.peermodelsnetwork.com/route/voi/run## $pars
## [1] "p1"
##
## $evppi
## [1] 0.0881INLA method
evppi.remote(outputs_nb, inputs, pars=c("p1","p2"), method="inla")Tuning the generalized additive model method
evppi.remote(outputs_nb, inputs, pars=c("p1","p2"), method="gam", gam_formula="s(p1) + s(p2)")## Calling server at https://prism.peermodelsnetwork.com/route/voi/run## $pars
## [1] "p1,p2"
##
## $evppi
## [1] 0.476
evppi.remote(outputs_nb, inputs, pars="p1", se=TRUE, B=100)## Calling server at https://prism.peermodelsnetwork.com/route/voi/run## $pars
## [1] "p1"
##
## $evppi
## [1] 0.0822
##
## $se
## [1] 0.0068Single-parameter methods
evppi.remote(outputs_nb, inputs, pars="p1", n.blocks=20, method="so")## Calling server at https://prism.peermodelsnetwork.com/route/voi/run## $pars
## [1] "p1"
##
## $evppi
## [1] 0.08
evppi.remote(outputs_nb, inputs, pars="p1", method="sal")## Calling server at https://prism.peermodelsnetwork.com/route/voi/run## $pars
## [1] "p1"
##
## $evppi
## [1] 0.0815Traditional Monte Carlo nested loop method
Model evaluation function
model_fn_nb <- function(p1, p2){
c(0, p1 - p2)
}Parameter simulation function
par_fn <- function(n){
data.frame(p1 = rnorm(n, 1, 1),
p2 = rnorm(n, 0, 2))
}
Invoking evppi_mc.remote
evppi_mc.remote(model_fn_nb, par_fn, pars="p1", ninner=1000, nouter=100)## Calling server at https://prism.peermodelsnetwork.com/route/voi/run## $evppi
## [1] -0.1702Accounting for parameter correlation
par_fn_corr <- function(n, p1=NULL){
p1_new <- if (is.null(p1)) rnorm(n, 1, 1) else p1
data.frame(p1 = p1_new,
p2 = rnorm(n, p1_new, 2))
}
evppi_mc.remote(model_fn_nb, par_fn_corr, pars="p1", ninner=1000, nouter=100)## Calling server at https://prism.peermodelsnetwork.com/route/voi/run## $evppi
## [1] -0.4713Expected value of sample information
Function to generate study data
datagen_normal <- function(inputs, n=100, sd=1){
data.frame(xbar = rnorm(n = nrow(inputs),
mean = inputs[,"p1"],
sd = sd / sqrt(n)))
}
set.seed(1)
evsi.remote(outputs_nb, inputs, datagen_fn = datagen_normal, n=c(10,100,1000))## Calling server at https://prism.peermodelsnetwork.com/route/voi/run## $n
## [1] 10 100 1000
##
## $evsi
## [1] 0.0723 0.0809 0.0817Built-in study designs
evsi.remote(outputs_nb, inputs, study = "normal_known", n=c(100,1000), pars = "p1")## Calling server at https://prism.peermodelsnetwork.com/route/voi/run## $n
## [1] 100 1000
##
## $evsi
## [1] 0.0804 0.0824
evsi.remote(outputs_cea, inputs, study = "normal_known", n=c(100,1000), pars = "p1")## Calling server at https://prism.peermodelsnetwork.com/route/voi/run## $n
## [1] 100 100 100 1000 1000 1000
##
## $k
## [1] 1 2 3 1 2 3
##
## $evsi
## [1] 0.0814 0.1686 0.2558 0.0813 0.1697 0.2580Importance sampling method
likelihood_normal <- function(Y, inputs, n=100, sig=1){
mu <- inputs[,"p1"]
dnorm(Y[,"xbar"], mu, sig/sqrt(n))
}
evsi.remote(outputs_nb, inputs, datagen_fn = datagen_normal, likelihood = likelihood_normal,
n=100, pars = "p1", method="is", nsim=1000)## Calling server at https://prism.peermodelsnetwork.com/route/voi/run## $n
## [1] 100
##
## $evsi
## [1] 0.0883
evsi.remote(outputs_nb, inputs, study = "normal_known", n=100, pars = "p1", method="is", nsim=1000)## Calling server at https://prism.peermodelsnetwork.com/route/voi/run## $n
## [1] 100
##
## $evsi
## [1] 0.0887Value of Information in models for estimation
p1 <- rbeta(10000, 5, 95)\[p_2 = expit(logit(p_1) + \beta)\].
beta <- rnorm(10000, 0.8, 0.4)
p2 <- plogis(qlogis(p1) + beta)
plot(density(p1), lwd=2, xlim=c(0,1), main="")
lines(density(p2), col="red", lwd=2)
legend("topright", col=c("black","red"), lwd=c(2,2),
legend=c("Surveyed infection probability", "True infection probability"))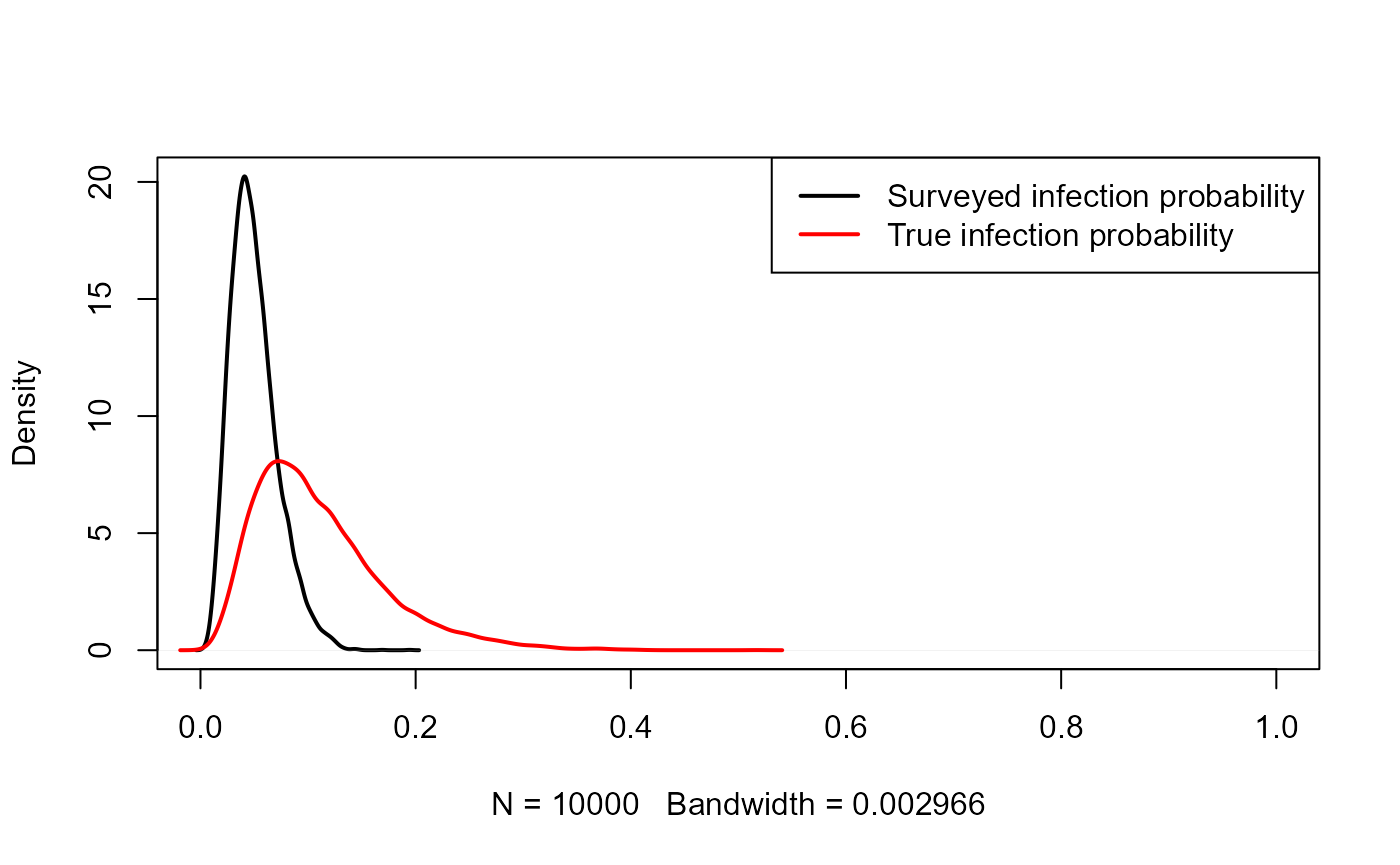
EVPI and EVPPI for estimation
var(p2)## [1] 0.0035409
inputs <- data.frame(p1, beta)
(evppi_beta <- evppivar.remote(p2, inputs, pars="beta"))## Calling server at https://prism.peermodelsnetwork.com/route/voi/run## $pars
## [1] "beta"
##
## $evppi
## [1] 0.0015
(evppi_p1 <- evppivar.remote(p2, inputs, pars="p1"))## Calling server at https://prism.peermodelsnetwork.com/route/voi/run## $pars
## [1] "p1"
##
## $evppi
## [1] 0.0019## [1] 0.05950546## [1] 0.04517632## [1] 0.04050802How regression-based EVPPI estimation works
plot(x=p1, y=p2, pch=".")
mod <- mgcv::gam(p2 ~ te(p1, bs="cr"))
p1fit <- fitted(mod)
lines(sort(p1), p1fit[order(p1)], col="blue")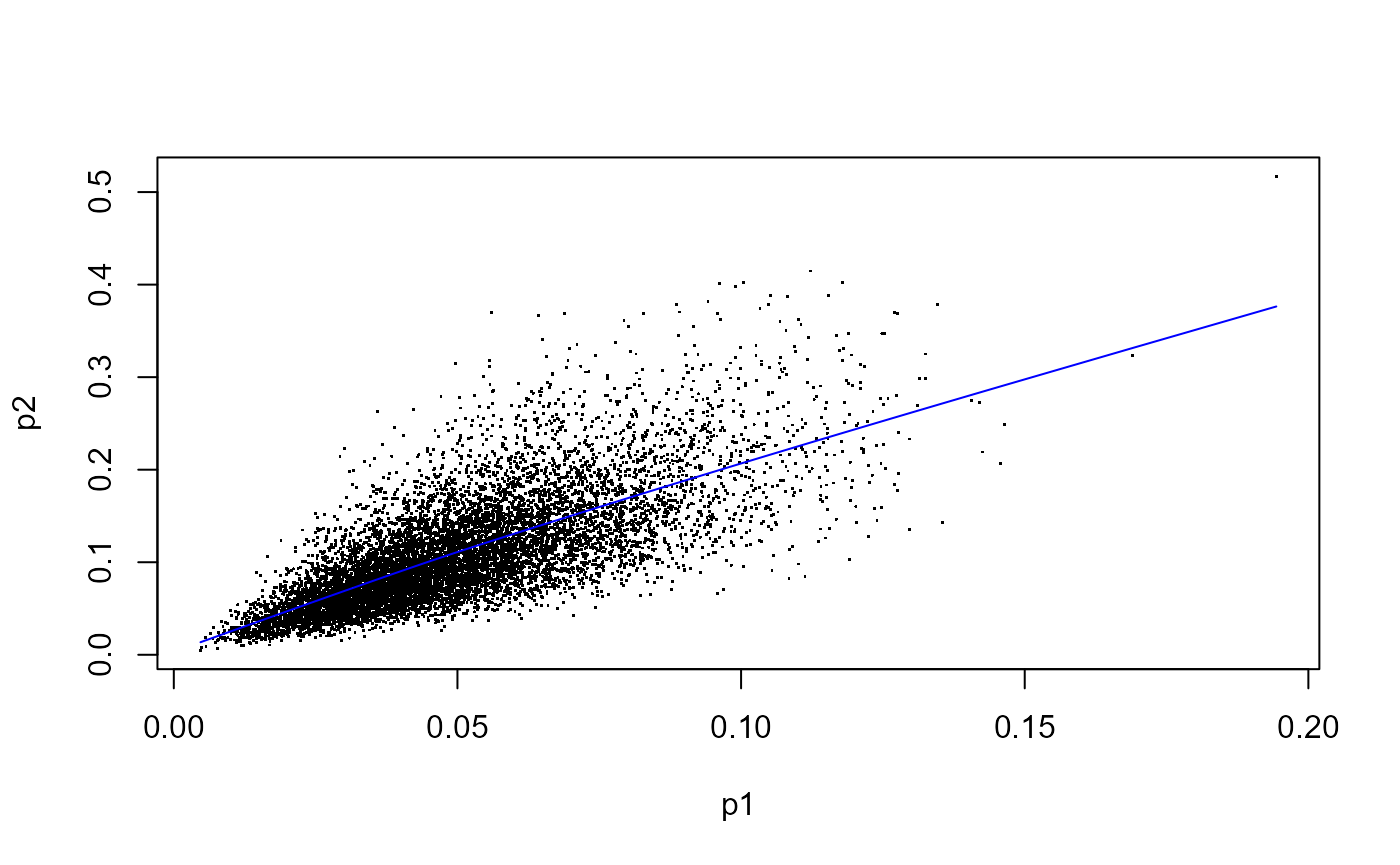
## [1] 0.001888821\[var(Y) - E_{X}\left[var_{Y| X}(Y |X)\right] = var_{X} \left[E_{Y|X}(Y|X)\right]\]
var(p1fit)## [1] 0.001887927EVSI for estimation
evsivar.remote(p2, inputs, study = "binary", pars="p1", n=c(100,1000,10000))## Calling server at https://prism.peermodelsnetwork.com/route/voi/run## $n
## [1] 100 1000 10000
##
## $evsi
## [1] 0.0010 0.0017 0.0019
inputs_p2 = data.frame(p2 = p2)
evsivar.remote(p2, inputs=inputs_p2, study = "binary", pars="p2", n=c(100, 1000, 10000))## Calling server at https://prism.peermodelsnetwork.com/route/voi/run## $n
## [1] 100 1000 10000
##
## $evsi
## [1] 0.0028 0.0035 0.0035